ran gel from 8/9 and got bars. at a loss.

august 9 2011
results from yesterdays water qPCRs are below this time got five out of six peaks.Set up 6 water PCRs to run tomarrow.


august 8 2011
Water samples had some amplification in two of the samples alomng the same places we have been seeing peaks up to this point. a picture of the melt curve can be seen HERE. set up qPCR run consisting of six 20 ul samples that are currently in the 4 degree fridge.
August 4 2011
Amplification and melt curve data from yesterdays run are below. Going to run just primers and water to see oif we still get a curve. Set up 4 qPCR samples with just water and put them on a tray in the 4 degree fridge.


August 3 2011
Ran an alighment of the primers against the CH-def H1 and 2. HERE are the results for primers 1070 and 1109 respectively. seems like the primers used may be amplifying multiple producs. also prepared an new PCR using half the previos amounts of primers and replacoing it with water.
August 2 2011
Looked up sequences for Cg def Cg defh1 and Cg defh2 they can be found HERE
july 25 2011
looking to start from from scratch and find new primers. did a blast search and came up with the following little table
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
|---|---|---|---|---|---|---|---|
| Forward primer |
TGCTGCTTTGGTGGCAGCCT |
Plus |
20 |
1269 |
1288 |
59.47 |
60.00% |
| Reverse primer |
TGCACCCCATTGTCCCCAAAG |
Minus |
21 |
1475 |
1455 |
57.18 |
57.14% |
| Product length |
207 |
||||||
July 21 2011
ran a qPCR with following amounts 198 ul of SSo, 20 ul of primers 180 ul of H2O. protocol was 98 for 2 min, 98 for 5 sec 60 for 5 sec.july 21 2011July 21 2011
got a amplfication chart and a melt curve. got two peaks so there might be dimer primer being replicated. will try it again at 65 degrees.


a copy of the report can be found HERE
July 15 2011
Ran gel with PCR products from 7/11. from left to right laddr, cleans 1-8, infected 7, 2, 1, 3, 4, 5, 6, 8 and controls. It is a litle wierd that in this one all of the clean samples show bars, not sure why that it is different here.

July 14 2011
Set up PCR using infected and control DNA. set up 16 samples and two controls. PCR parameters:
95 C for ten min
95 C for 15 sec
65 C for 15 sec
72 C for 30 sec
July 11 2011
Ran gel from 6.28. from right to left ladder infected 1-8 and controls 1-8. there appeared to be some dimer primer but gene was expressed in all diseased oysyers and only about half of the controls.
June 28 2011
Set up PCR for sixteen samples using 220 ul of master mix, 20 ul of mixed forward and reverse primers 1070 and 1109, and 185 ul of H2O. for the templates used DNA extracted from oysters used in an experiment on 1/11/11. There were 8 samples from oysters infected by vibro and 8 controls taken at hour one. Infected samples are in purple PCR tubes and controls are in blue. PCR parameters are as follows:
95 C for ten min
95 C for 15 sec
55 C for 15 sec
72 C for 30 sec
is programed into the PCR machine as AAAAA in sams folder.
Ran the PCRs from the twentieth and they WORKED!! PERFECTLY!! left to right ladder experiment experiment control control.
June 20 2011
Ran out gels from 5/26 and 6/2 having picture uploadinjg issues but the 5/26 had nothing and the ones from 6/2 a bar but also had primer dimer. I am going to use the primers that derrek so thoughfully fojnd that work and hopefully can soon begen to experiment. also used the last of the 1x TAE so made more by mixing 40 mils of 25x TAE with 960 mills of nanopure H2O.
june 2 2011
Ran a PCR using the following parameters using the f and r primers from 1/20/11. also used a new cDNA of C gigas gil from BB pool55 ul of Master mix, 46.2 ul of H2O, 2.2 ul of primers and 1 ul of template. Ran a PCR of 40 replications of the following parameters:
95 C for ten min
95 C for 15 sec
55 C for 15 sec
72 C for 30 sec
may 26 2011
Ran a PCR using the following parameters both forward and reverse were mixed in one tube. their SR ids are 1109 and 1070. also used a new cDNA of C gigas gil from BB pool55 ul of Master mix, 46.2 ul of H2O, 2.2 ul of primers and 1 ul of template. Ran a PCR of 40 replications of the following parameters:
95 C for ten min
95 C for 15 sec
55 C for 15 sec
72 C for 30 sec
may 6 2011
set up a PCR gopefully going to use other machine. also runinng hel from yesterdays PCR. One of the experimental ones evaporated so I am only running one experiment one gel and THE LADDER!!!
ran it out and got results. going to do one more on monday and if it works figure out what the next steps are.
ladder experiment control
Cinco de Mayo 2011
Ran a PCR using the following parameters both forward and reverse were mixed in one tube. their SR ids are 1109 and 1070. also used a new cDNA of C gigas gil from BB pool55 ul of Master mix, 46.2 ul of H2O, 2.2 ul of primers and 1 ul of template. Ran a PCR of 40 replications of the following parameters:
95 C for ten min
95 C for 15 sec
55 C for 15 sec
72 C for 30sec
got what appeared to be bands but no ladder so going to run it again tomarrow fingures crossed and for some reson i can not get the picture off of sites:( but its there
Ran another PCR with same parameters as above.
May 2 2011
got new primers. both forward and reverse were mixed in one tube. their SR ids are 1109 and 1070. set up a pcr with the following parameters: 55 ul of Master mix, 46.2 ul of H2O, 2.2 ul of primers and 1 ul of template. Ran a PCR of 40 replications of the following parameters:
95 C for ten min
95 C for 15 sec
60 C for 15 sec
72 C for 30 sec
going to run it out on one percent gel made with 75 ul of TAE buffer .75 g of agarose and 6 ul of ethdium bromide.
nothing showed up on the gel. I couldnt find the camera to take a picture.
April 8 2011
got a new Def primer and made 2 runs of 4 samples using the following parameters 55 ul of Master mix, 46.2 ul of H2O, 2.2 ul of primers and 1 ul of template. Ran a PCR of 40 replications of the following parameters:
95 C for ten min
95 C for 15 sec
60 C for 15 sec
72 C for 30sec
Non of them worked so going to retry sunday using new genom,ic/cDNA templates
from left to right ladder old primer cDNA old primer cDNA control control new primer cDNA new primer cDNA control control

April 7 2011
prepared for four runs using 55 ul of Master mix, 46.2 ul of H2O, 2.2 ul of primers and 1 ul of template. Ran a PCR of 40 replications of the following parameters:
95 C for ten min
95 C for 15 sec
60 C for 15 sec
72 C for 30sec
had some evaporation issues again. I thought the caps seemed a little loose but I just pushed down on them really hard and hoped for the best. Ill be more careful about it next time. failed again picyure below cDNA, cDNA, control control. going to try using new primers tomorrow.

April 3 2011
ran a PCR and ran two gels using following parameters and got zilch.
prepared for four runs using 55 ul of Master mix, 46.2 ul of H2O, 2.2 ul of primers and 1 ul of template. Ran a PCR of 40 replications of the following parameters:
95 C for ten min
95 C for 15 sec
60 C for 15 sec
72 C for 15 sec
April 1 2011
trying to get some results with the gigas cDNA again. prepared for four runs using 55 ul of Master mix, 46.2 ul of H2O, 2.2 ul of primers and 1 ul of template. Ran a PCR of 40 replications of the following parameters:
95 C for ten min
95 C for 15 sec
60 C for 15 sec
72 C for 15 sec
Ran gel from March 31, from left to right ladder cDNA, cDNA control control, got a proper sized bar in the first experiment but is a little smuged try again monday

March 31 2011
Back for more abuse:). trying to get some results with the gigas cDNA again. prepared for four runs using 55 ul of Master mix, 46.2 ul of H2O, 2.2 ul of primers and 1 ul of template. Ran a PCR of 40 replications of the following parameters:
95 C for ten min
95 C for 15 sec
60 C for 15 sec
72 C for 15 sec
was going to try to run out on a plate as well but all the electrolysis stuff was in use, so be back tommara:)
Feburay 15 2010
Ran PCR products out on gel. From left to right: ladder cDANA, cDNA, control, control, DNA, control control. I set up a run of just genomic DNA with the following specs 55 ul of master mix, 46.2 ul of water and 4.4 ul of each primer. this was ging to be used to make four runs, two with DNA template ans two as controls. 1 ul of gigas gill genomic DNA was added to two of the tubes and one ul of water was added to the other two as controls
Parameters for PCR
95 C for ten minutes
95 C for 15 sec
55 C for 15 sec
72 C for 30 sec
repeated 40 times
Held at 72 c for ten min

Feburay 10 2011
set up and ran PCR of DNA and cDNA with all the same specs as the other times
Feburary 8 2011
another epic failure, from left to right: ladder, cDNA, cDNA control control DNA DNA control control. going to try again
Feburary 3 2011
Running out PCR cDNA product on a gel. from ledft to right: ladder cDNA, cDNA, control, control, DNA, DNA, control, control
Ran out PCR product at an annealing temperature of 60 and got a band. going to sequence it to see if it is the right one. Set up a second PCR with both cDNA templates and DNs templates (sepretly) using the same primer
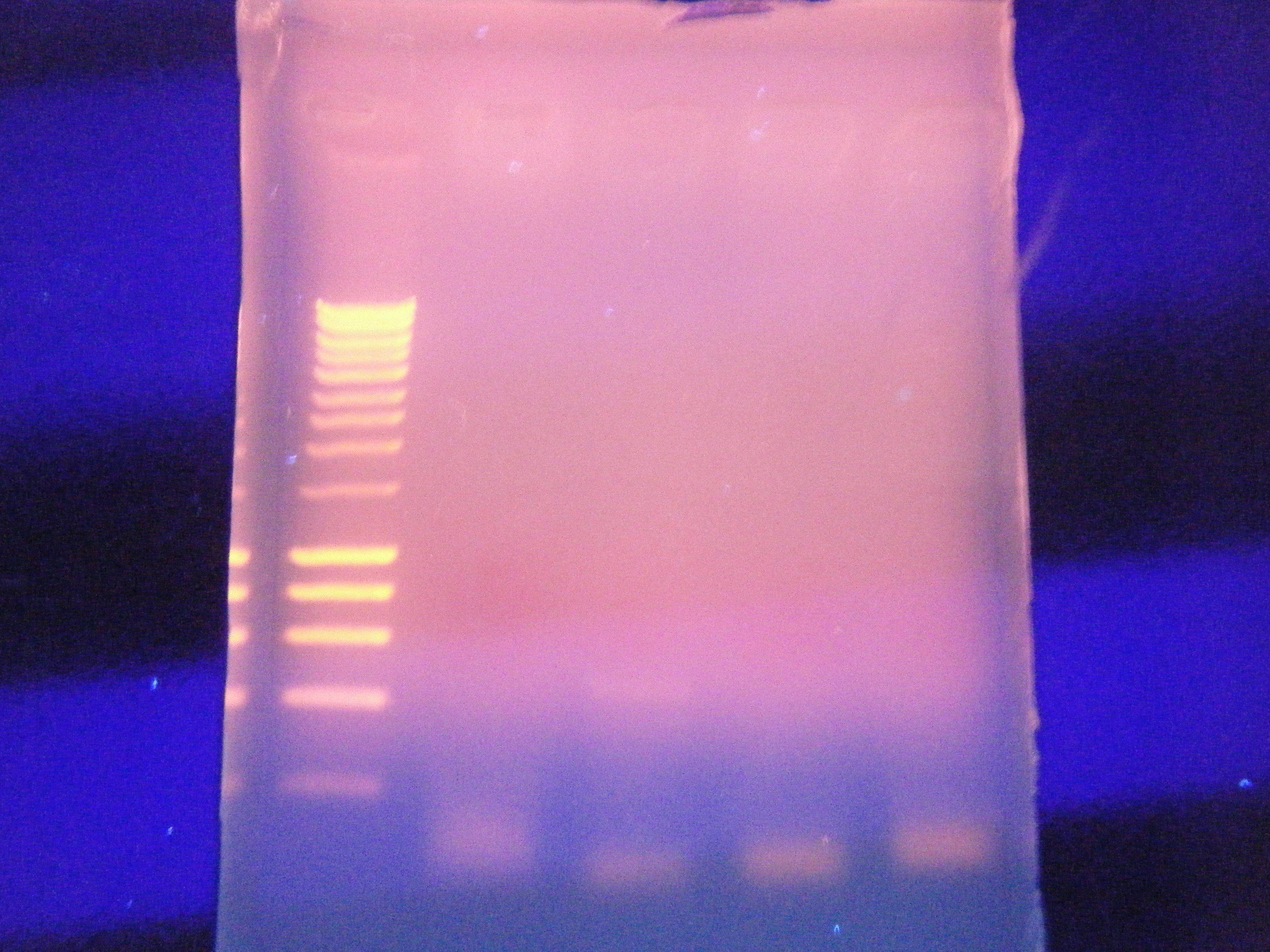
From left to right: ladder, cDNA, cDNA, control contro
January 26 2011
Ran PCR product that was at 63 degrees and got no bands. the temperature was probably to high. going to try again with an annealing temperature of sixty and see if that works.
made gel and running out the PCR products on it. Got two distinct in the experiment. going to up the annealing temperature to 63 and try again
January 20 2011
Testing primers in a PCR
using pool c gigas cDNA. Making enough for four runs, two with template and two control. added aditional 10 percent to each reaction
Master mix 55 ul
F primer 2.2 ul
R primer 2.2 ul
h20 46.2 ul
1 ul of cDNA each
PCR parameters
95 C for 10 min
95 C for 10 sec
55 for 15 sec
72 for 30 sec
forty repeats
January 10 2011
Found the primers needed to copy C. gigas defensin mRNA. In order to find them I searched for the oyster defensin protein and looked at related sequences and found Crassostrea gigas isolate H1-B215 defensin (defs) mRNA, complete cds. I ran a primer blast on it and got the following
||
| Sequence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
|
|---|---|---|---|---|---|---|---|
| Forward primer |
ACACTGGTCGTTCTCCTGATGGT |
Plus |
23 |
22 |
44 |
57.44 |
52.17% |
| Reverse primer |
ACTGGTCACGCGGACACCCA |
Minus |
20 |
91 |
72 |
60.39 |
65.00% |
| Internal oligo |
Plus |
||||||
| Product length |
70 |
||||||
September 23 2009
Set up PCR of spot fan 3 with 10x, 100x and 1000x dilutions, everything same as previous runs.
September 22 2009
Heated spot fan samples at 95 C for 20 min spun in 4C for 15 and put supernatant in new container. put samples through nanodrop, results below (ignore any second duplicate)

September 21 2009
Ran gel of diluted DNA got some results going to think about what to try next.
From right to left Spot fan 2 diluted 10x, 100x, 1000x, control, control, control

September 19 2009
Did some computer editing work. Set up a PCR runs of spot fan 2, serially diluted 10, 100 and 1000 times and EUK a/b primers. When diluting I used 3 ul of DNA and 27 ul of PCR H2O, in retrospect I only need 2 ul of DNA, so I could have gotten away with much smaller amounts. other than that the run is the same as the last couple of runs, but hopefully we will get some useable results this time.
September 18 2009
Did some editing of spreadsheets and updating the power point. ran gel of LABY primer dea fan and got similar results. Apparently the same sort of thing was happening a year ago when sea fan was being worked with here.
From right to left: spot fan 2, 3, 9, 10 and control
September 16 2009
Did some spreadsheet updating finding the primers used in trout experiment. Set up new PCR of sea fan DNA with spot fan samples 2, 3, 9, 10, this time using LABY primer, Used 25 ul of master mix, 21 H2O, 1 of primer and 2 DNA except for the negative control (which I remembered this time:) that had an additional 2 ul of H2O.
Parameters of PCR run:
94 for 30
50 for 30
72 for 90
Repeated 39 times
edited spreadsheet.
September 15 2009
Compiled charts into single power point.
September 14 2009
Ran gel of remaining sea fan DNA and got more smears, ran pure DNA and got nothing. Probably going to have to do this over from scratch.
From left to right: spot fan 2,3, 9,10,11, 12, 13, 14, 15, control fan 1, 4, 5, 6, 7, 8
September 11 2009
collected and aggregated algae growth data into a chart, see below, also ran a gel on one of the sea fan samples, also see below. Remember next time to run a control PCR!!!!
From left to right: Sea fan 1,4,5,6,7,8

September 10 2009
Chelaxed samples of control and regular sea fan pictured below. Set up a PCR, used 25 ul of Master mix, 21 of H2O, 1 of primers EUK A and B, and 2 ul of DNA.
PCR Perameters
94 for 30 sec
50 for 30 sec
72 for 90 sec
September 9 2009
Chelaxed samples of diseased sea fan, pictured below.
September 8 2009
Added 25 Tw 1052 to 500 mil media that was prepared on the 3rd. added fresh media to older stock. Ts are at 104000
September 4 2009
Set up media for TW 1052 using the same method as for the TS except added 6 mg of sodium meta silcate. TSs still growing but slowing down at 106667 per mil.
September 3 2009
the TWs are still steady and the TSs are starting to level of, today they were at 104533. Finished analyzing Macs data.
september 2 2009
The TW 1052 remained constant but the Ts went up to 103200 per mil. Basically finished enterinig Macs data, have a little analysis left to do.
September 1 2009
Algae counts were basically identical to yesterday, I'll count for two more days before deciding if this is the max. Also entered a bunch of data for Mac, will probably finish it off tomorrow.
August 31 2009
The TS and TW 1052 more then doubled their populations over the weekend with populations of 100000 and 71000 per mil respectively. On a saddr note the tw1336 is pretty much extinct. today's pH was 7.16.
August 28 2009
Took 25 mils from the Terta Selmis separated on 8/4 and added them to the freshly autoclaved 500 mills of media. Also added 25 mills of fresh media to the older flask. the pH is 6.9. the counts of the algae are as follows (corrected for dilution) the TW 1052 are still hovering around 48000 a mil while the TS jumped up to 856000.
August 26 2009
Made media for Tetra selmi. used .5 liters of fiultered sea water. Instuctions on the bottles of nutrieants called for 10 mils per 20 gallons and half a liter is .132806 gallons so I used .5*.132806 mils of the two soulutions which came to 66 ul. The algea counts corrected for dilution are as follows TS: 53867 TW 1052 48000 and TW 1336 10000 per mil. will see tommaroow if they continue to go down. the ph was 7.1.
August 25 2009
The observation I made yesterday about the water temp and pH seems to be correct, I made sure to let the mater get to room temp today and got a reading very similar to yesterday, 7.4. the count for the TS is 109867 per mil, tW 1052 58700 and the TW 1336 is at 44300.
August 24 2009
Started count of algae separated out on 8/17. Corrected for dilution I got the following: TW 1336 41800 per mil TS 82667.TW 1052 53400 per mil. The pH for the CO2 tank read 7.5 today, this might be because I made sure to let the sample get to room temp, I will do the same for the rest of this week to make sure.
August 18 2009
The new TWs I set up yesterday seem to be doing pretty well, there are 56400 TW 1336 per mil and 25000 TW 1052 per mil, the TS have reached a point were there are over 330000 per mil so I took out 10 mils of it and started a new batch. the CO2 level in tank A is 6.9
August 17 2009
Set up spread sheets and graphs of algae counts and ph levels. the can be seen at http://spreadsheets.google.com/ccc?key=0Aol6idbLnLITdGRuczVGOXZsUFAzUGRRdHBWVGNXb3c&hl=en and http://spreadsheets.google.com/ccc?key=0Aol6idbLnLITdGhDQnpXQlhxLVVzelFxUHd4LUtPNHc&hl=en respectively. All the TS's I set up earlier have died out do I am going to set up another sample from scratch.
August 13 2009
The pH reader downstairs musty be a little off because I am getting readings of 6.8-6.for the CO2 tank and it is reading 7.2 downstairs. The control is at a steady 8. The TW algae I split on 8/4 seem to be pretty much decimated they are in the low tens per micro liter and there are a lot of dead cells floating around. I split off a 10 mil sample from the 8/4 split and missed it with 40 mils of media so we will see if it happens again. The TS are doing great and their concentration adjusted for dilution is 178133 per mil.
August 12 2009
Co2 tank is reading a pH of 6.9, the sensor downstairs says 7.2. the control tub is a healthy 8.
counted the newer algae samples. the following are the concentration per mil adjusted for dilution TW 1052: 18200 TW 1336: 19600 TS:142700
August 11 2009
When collecting water from the CO2 tank noticed that the CO2 bubbler was not running. this could explain why there was still a relatively high pH yesterday. So took new pH readings after the CO2 had been running for a while and got 7.6 for tank A and 8 for both of the tanks had a SG of 1.024 and a temp of 9 C.
I counted the algae using the same method as before and have come to the following concentration ber mil, accounting for dilution: TW 1336:137800 per mil, TW 1052: 141800 TS: 206400
August 10 2009
Salinity, temperature, and pH of downstairs oyster tanks were all similar to the readings that were taken on 8/5 except for the CO2 tank which is at 44.
Counted algae using same methods as on 8/5, averages were as follows, tw 1336: 97 per ul tw 1052: 92 TS: 146. the volumes per ml corrected for dilution are 97000, 92000 and 195000 respectively. Also some how the machine that held the TWs heater was turned off this weekend and temperatures dropped a slow as .4 C, this could account for a larger amount of dead cells in those samples.
August 5 2009
Took Salintiy and pH, however this time when I took the pH I span the sample and got a pH of 7.88 for the take with CO2 (A) and a pH of 8.1 for the control (b). Both tanks have a SP of 1.024 and a temperature of 9 C. The pressure in the CO2 tank is currently at 56 kg/cm2.
The average for the TW 1052 alge was 66, the TW 1336 was 72 and the TS was 155. for the TWs my sample size was 50 ul and for the TS it was 60 ul and 20 ul of 70% ethonal. THe averages where calculated from counting the toatal in 1 ul of the solution. with the above numbers in mind we can conclude that there are about 66000 TW 1052, 72000 TW 1336, and 207000 TS per mil. The data can be seen at http://spreadsheets.google.com/ccc?key=0Aol6idbLnLITdGRuczVGOXZsUFAzUGRRdHBWVGNXb3c&hl=e
August 4 2009
Both tubs A and B had pHs of about 7.76, temperatures of 9 C and had a SG of 1.024.
Counted How many T Selmis where in five random squares
1.175
2. 120
3. 200
4. 224
5. 250
TW 1336 TW 1052
1. 61 1. 61
2. 51 2. 48
3. 59 3. 49
4. 65 4. 40
5. 62 5. 59
Made new cultures of the T Selmis and the TW algae.
Added 10 mil of the algae to 40 mils of the media in a 250 mil flask. The TW is at 16 degrees and the T Selmi is at RT.
July 29 2009
Oysters Downstairs are all still healthy in water that is a crisp 9C with a specific gravity of 1.024.
Ran a gel of the algal DNA I PCRed on the 28th. much better:D despite some odd inconsistencies:(
From the left: ladder, control, control, DNA DNA

July 28 2009
Ran second PCR on algal DNA. Basically the same as on 7/2709 except use EUK A and EUK B primers instead of the ones I ordered.
Finished running gel, failed miserably:( on the other hand the ladder turned out OK :P

ran electrolysis
(From left to right)
cell 1 is ladder
cells 2 and 3 are experiment
4 and 5 are controls
ALWAYS RUN NEGATIVE TO POSITIVE!!!
Set up electrolysis gel (small), USE GLOVES!
1. 75 ml of TAE buffer
2. 1 g agar
3. heat for 3 min, mixing every min
4. add 6 ul of ethdium bromide (fridge)
5. pour into plate, rest for 35 min
July 27 2000
Set up a PCR run for algal DNA
25 ul Master mix
1 ul of primers
2 ul of DNA
21 ul of H20
I made to MM for all four runs at the same time to minimize pipetter error.
Reverse transcripted RNA of oyster, accidentally made second experiment instead of control:(
1. heat RNA for 5 min at 75
2. make master mix
4 ul of buffer
8 ul DNTP
1 ul Reverse transcriptase
1 ul dt primer
1 ul RNAse free H2O
3. mix MM and rna
4. vortex
5. spin and rest at RT for 10 min
6. Incubate at 75 for 1 hour
7. heat inactivate 95 C for 3 min
8. store at -20
July 24 2009
Extracted RNA from Littorina pictured below
and got the following Nano drop results

July 22 2009
Extracted RNA from barnacles, got to much tissue so I had to divide it into 4 different tubes, and consolidated the RNA after the resuspension. Mass speced.

July 21 2009
Extracted RNA From a giant pacific Oyster
1.Place sample (cutting of oyster gill) in tube
2. add 500 micro liters of tri reagent
3. homogenize
4. add 500 micro liters of tri reagent and rest at RT for 5 min
5. add 200 micro liters of chloroform
6. rest at RT for 10 min
7.centrifuge at 12000 g and 4 C for 15 min
8. remove supernatant
9. add 500 micro liters of isopropanol to supernatant.
10. rest for 10
11centerfuge at 12000 g and 4 C for 8 min
12. remove supernatant
13 add 1 ml of 75 percent ethanol
14. centrifuge at 7500 g for 5 min
15 remove supernatant
16. add 50 micro liters of treated h20
17. heat at 50 C for 10 min

Oyster stats
| CO2 treated Oysters |
length (mm) |
Weight (g) |
| A |
147 |
188.9 |
| b |
145 |
207.1 |
| c |
61 |
283.19 |
| d |
94 |
283.19 |
| e |
103 |
187.9 |
| f |
100 |
187.9 |
| Control Oysters |
||
| 1 |
105 |
176.3 |
| 2 |
137 |
229 |
| 3 |
98 |
138.06 |
| 4 |
90 |
129.13 |
| 5 |
133 |
196.22 |
| 6 |
121 |
179.11 |
July 20 2009
Need to order primer for PCR run of EU555177 gene of algae. this gene has to do with the alges ability to absrobe light. primer chosen is:
| ence (5'->3') |
Strand on template |
Length |
Start |
Stop |
Tm |
GC% |
||
|---|---|---|---|---|---|---|---|---|
| Forward primer |
ACACAATTGGAATGCGAACA |
Plus |
20 |
101 |
120 |
59.97 |
40.00% |
|
| Reverse primer |
GCTAAATGGTGGTGAGCCAT |
Minus |
20 |
563 |
544 |
59.96 |
50.00% |
|
| I |
||||||||
July 17 2009
Set up instant ocean baths with a gravity of 1.024 for ocean acidification experiment.
Chelaxed samples of copepods and seaweed.\
Chellaxing:
1. If alive kill, otherwise just centrifuge.
2. Add 250 micro liters of Chelax (found in fridge of lab 1), shake chelax before using.
3. vortex sample
4. Heat for half an hour min
5. vortex